Molecular Investigation of gyrA Mutations in Clinical Isolates of Methicillin-Resistant Staphylococcus aureus Derived from Diverse Sources
DOI:
https://doi.org/10.54133/ajms.v5i1S.282Keywords:
Antibiotics, Fluoroquinolone, gyrA, Mutations, Staphylococcus aureusAbstract
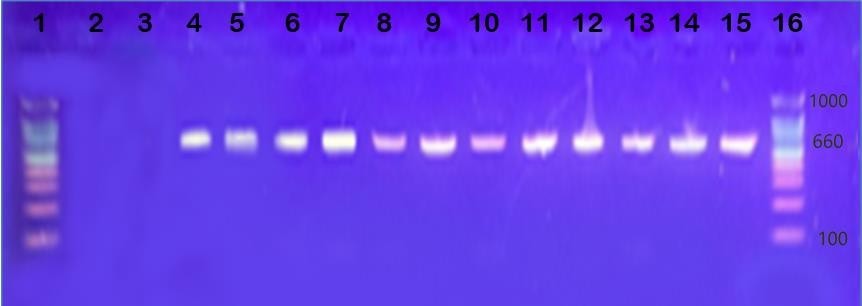
Background: Fluoroquinolones are the most effective antibiotics against Staphylococcus aureus isolates. In hospitals, excessive use of antibiotics has led to the emergence of highly resistant strains of S. aureus isolates. Objective: The aim of this study was to detect the mutations that occur in the gyrA gene encoding for DNA gyrase, which is one of the targets for fluoroquinolone resistance. Methods: Fifty clinical isolates were diagnosed as S. aureus according to molecular and bacteriological methods. The susceptibility tests were performed on all bacterial isolates by the disc diffusion method using methicillin and six fluoroquinolone antibiotics. Results: Out of fifty isolates, twelve were resistant to methicillin and all six antibiotics (nalidixic acid, lomefloxacin, ciprofloxacin, norfloxacin, ofloxacin, and levofloxacin). From the fifty isolates, 12 were resistant, 3 were intermediate, and 38 were sensitive to three or more tested antibiotics. The resistance of S. aureus isolates was also confirmed by the minimum inhibitory concentration test. The main sources of isolates were burns (10%), nose (16) wounds (8%), operation room (10%), ear (20%), urine (8%), skin (6%), and throat (22%). Twelve resistant isolates were used to examine the mutations in the gyrA gene. A direct sequence analysis found eight mutations in the gyrA gene; these mutations included 2 (25% missense mutations), 1 (12.5%) deletion mutation, and 5 (62.5%) silent mutations at various sites. Conclusion: gyrA mutations resulting from the excessive use of antibiotics may be one of the mechanisms leading to fluoroquinolone resistance.
Downloads
References
Chinnambedu RS, Marimuthu RR, Sunil SS, Amrose P, Ramachandran V, Pachamuthu B. Changing antibiotic resistance profile of Staphylococcus aureus isolated from HIV patients (2012–2017) in Southern India. J Infect Public Health. 2020;13(1):75-79. doi: 10.1016/j.jiph.2019.06.015. DOI: https://doi.org/10.1016/j.jiph.2019.06.015
Rana A, Kumar NR, Kaur J. Therapeutic effect of propolis on Staphylococcus aureus induced oxidative stress in spleen of Balb/c mice: A biochemical and histopathological study. Indian J Nat Prod Resource. 2022;13(3):346-355. doi: 10.56042/ijeb.v60i08.54823. DOI: https://doi.org/10.56042/ijeb.v60i08.54823
Maree M, Thi Nguyen LT, Ohniwa RL, Higashide M, Msadek T, Morikawa K. Natural transformation allows transfer of SCCmec-mediated methicillin resistance in Staphylococcus aureus biofilms. Nat Commun. 2022;13(1):2477. doi: 10.1038/s41467-022-29877-2. DOI: https://doi.org/10.1038/s41467-022-29877-2
de Morais Oliveira-Tintino CD, Muniz DF, Dos Santos Barbosa CR, Silva Pereira RL, Begnini IM, Rebelo RA, et al. NorA, Tet(K), MepA, and MsrA efflux pumps in Staphylococcus aureus, their inhibitors and 1,8-naphthyridine sulfonamides. Curr Pharm Des. 2023;29(5):323-355. doi: 10.2174/1381612829666221212101501. DOI: https://doi.org/10.2174/1381612829666221212101501
Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone Antibiotics. 2019;10(10):1719–1739. doi: 10.1039/c9md00120d. DOI: https://doi.org/10.1039/C9MD00120D
Bush NG, Diez-Santos I, Abbott LR, Maxwell A. Quinolones: Mechanism, lethality and their contributions to antibiotic resistance. Molecules. 2020;25(23):5662. doi: 10.3390/molecules25235662. DOI: https://doi.org/10.3390/molecules25235662
Das B, Verma J, Kumar P, Ghosh A, Ramamurthy T. Antibiotic resistance in Vibrio cholerae: Understanding the ecology of resistance genes and mechanisms. Vaccine. 2020;38 Suppl 1:A83-A92. doi: 10.1016/j.vaccine.2019.06.031. DOI: https://doi.org/10.1016/j.vaccine.2019.06.031
Abd FB. Characterization of multidrud resistant Staphylococcus aureus isolated from various sources. Univers J Res Rev Arch. 2022;1(2):94-101.
Mhana SMY, Aljanaby AAJ. Bacteriological Investigation of Pathogenic Bacteria Causing Urinary Tract Infections: A cross-Sectional Study. IOP Conf Ser: Earth Environ Sci. 2023;1215:012067. doi 10.1088/1755-1315/1215/1/012067. DOI: https://doi.org/10.1088/1755-1315/1215/1/012067
Abd Al-Mayali M, Salman ED. Bacteriological and Molecular Study of Fluoroquinolones Resistance in Pseudomonas aeruginosa Isolated From Different Clinical Sources. Iraqi J Sci. 2020;61(9):2204-2214. doi 10.24996/ijs.2020.61.9.7 DOI: https://doi.org/10.24996/ijs.2020.61.9.7
Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol. 2021;59(12):e0021321. doi: 10.1128/JCM.00213-21. DOI: https://doi.org/10.1128/JCM.00213-21
Qamar S, Shaheen N, Shakoor S, Farooqi J, Jabeen K, Hasan R. Frequency of colistin and fosfomycin resistance in carbapenem-resistant Enterobacteriaceae from a tertiary care hospital in Karachi. Infect Drug Resist. 2017;10:231-236. doi 10.2147/IDR.S136777 DOI: https://doi.org/10.2147/IDR.S136777
Turki TG, Hashim NM, Abed BA, Al-Shuhaib MBS. Association of the growth hormone releasing hormone (rs368475481) polymorphism with acromegaly disorder in Iraqi patient. J N Z Herpetol. 2023;12:3112-3122.
Orfali RL, Silva Oliveira D, Lima D. Staphylococcus aureus enterotoxins modulate IL-22-secreting cells in adults with atopic dermatitis. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-25125-0. DOI: https://doi.org/10.1038/s41598-018-25125-0
Sanfilippo CM, Hesje CK, Haas W, Morris TW. Topoisomerase mutations that are associated with high-level resistance to earlier fluoroquinolones in Staphylococcus aureus have less effect on the antibacterial activity of besifloxacin. Chemotherapy. 2012;57(5):363-71. doi 10.1159/000330858 DOI: https://doi.org/10.1159/000330858
Ahmadi SH, Tabatabaie T, Ramavandi B, Hashemi SE. Amoxicillin enzymatic decomposition via H2O2-stimulation of catalase-positive bacteria in a sequential batch reactor. Environ Technol Innov. 2023;32:103352. doi: 10.1016/j.eti.2023.103352 DOI: https://doi.org/10.1016/j.eti.2023.103352
Kadhum HH, Abood ZH. Staphylococcus aureus incidence in some patients with atopic dermatitis in Baghdad city. Iraqi J Biotechnol. 2022;21(2):13-20.
Parmar AS, Panwar AS. Antimicrobial Study of Chitosan-Based Crosslinked Hydrogel Against Staphylococcus aureus, Porphyromonas gingivalis, Pseudomonas aeruginosa, and Streptococcus mutans. Current Trends in Biotechnology and Pharmacy. 2023;17(3s):1170-83. doi: 10.5530/ctbp.2023.3s.54
Al-Marjani MF, Kadhim KA, Kadhim AA, Kinani A. Ciprofloxacin Resistance in Staphylococcus aureus and Pseudomonas aeruginosa Isolated from Patients in Baghdad. Int Int J Pharm Sci Res. 2015;6(2):382–385.
Al-Jebouri MM, Mdish SA. Antibiotic resistance pattern of bacteria isolated from patients of urinary tract infections in Iraq. Open J Urol. 2013;03(02):124–131. doi: 10.4236/oju.2013.32024. DOI: https://doi.org/10.4236/oju.2013.32024
Rong D, Wu Q, Xu M, Zhang J, Yu S. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of staphylococcus aureus from retail aquatic products in China. Front Microbiol. 2017;8:714. doi: 10.3389/fmicb.2017.00714. DOI: https://doi.org/10.3389/fmicb.2017.00714
El-Tawab A, Ashraf A, Hofy FI, Mohamed SR, Amin SH. Characterization of methicillin resistance staphylococcus aureus isolated from chicken and human. Benha Vet Med J. 2017;32(1):132–137. DOI: https://doi.org/10.21608/bvmj.2017.31198
Khaleel D, Othman R, Khudaier B. Plasmid transformation and curing of nalidixic acid gene in Staphylococcus aureus isolated from buffaloes mastitis and workerʼs hands. Iraqi J Vet Sci. 2019;32(2):167–74. doi: 10.33899/ijvs.2019.153845. DOI: https://doi.org/10.33899/ijvs.2019.153845
Hashem RA, Yassin AS, Zedan HH, Amin MA. Fluoroquinolone resistant mechanisms in methicillin-resistant Staphylococcus aureus clinical isolates in Cairo, Egypt. J Infect Dev Ctries. 2013;7(11):796–803. doi: 10.3855/jidc.3105. DOI: https://doi.org/10.3855/jidc.3105
McCurdy S, Lawrence L, Quintas M, Woosley L, Flamm R, Tseng C, et al. In vitro activity of delafloxacin and microbiological response against fluoroquinolone-susceptible and nonsusceptible Staphylococcus aureus isolates from two phase 3 studies of acute bacterial skin and skin structure infections. Antimicrob Agents Chemother. 2017;61(9). doi.org/10.1128/AAC.00772-17. DOI: https://doi.org/10.1128/AAC.00772-17
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Al-Rafidain Journal of Medical Sciences ( ISSN 2789-3219 )

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Published by Al-Rafidain University College. This is an open access journal issued under the CC BY-NC-SA 4.0 license (https://creativecommons.org/licenses/by-nc-sa/4.0/).











