Dual Role of miR-150 in Colorectal Cancer Progression: A Quantitative Real-Time PCR Study
DOI:
https://doi.org/10.54133/ajms.v8i1.1757Keywords:
Colorectal cancer, Gene expression, miR-150-3p, miR-150-5p, MicroRNA, Pearson correlationAbstract
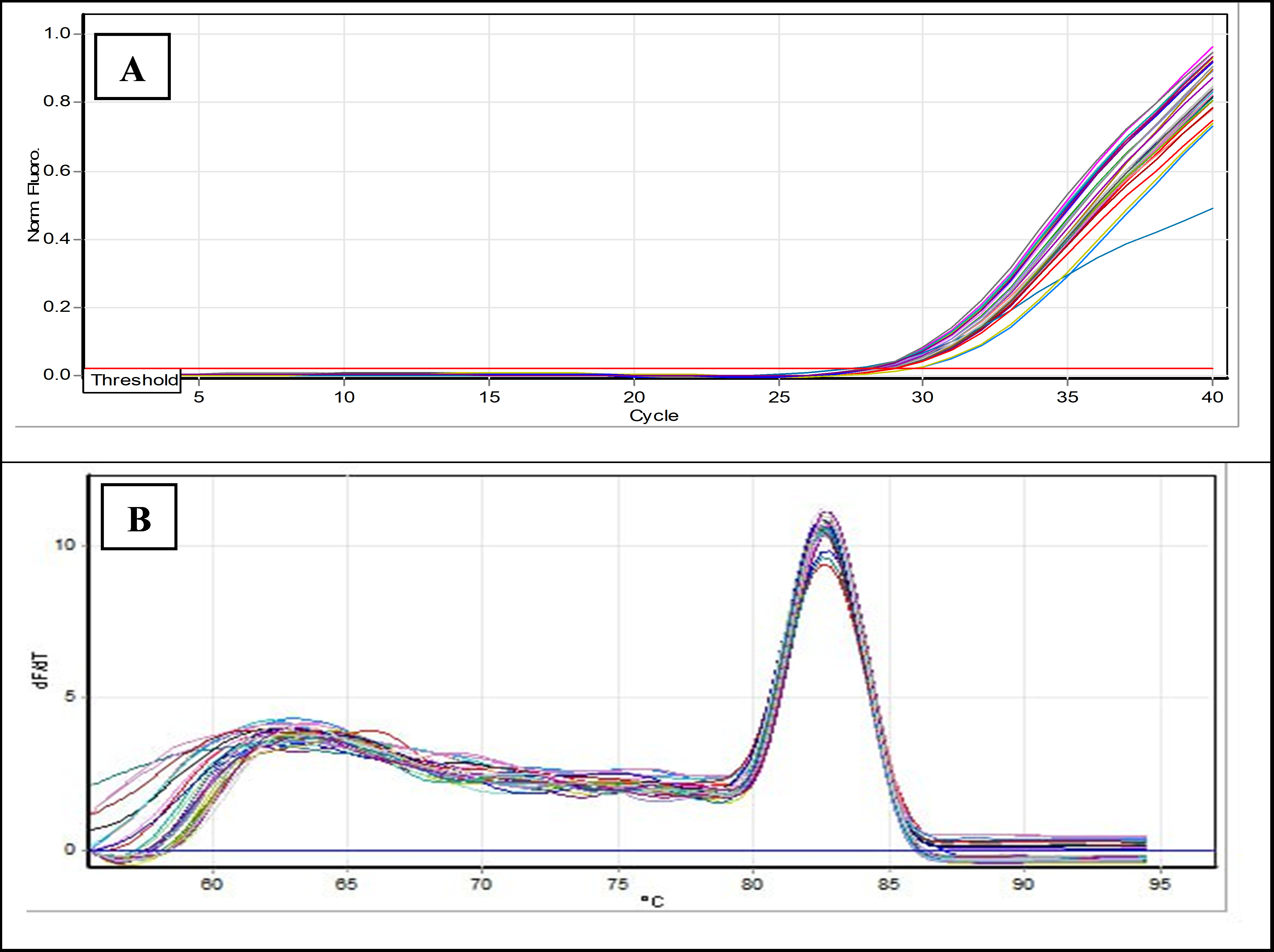
Background: Colorectal cancer (CRC) is the second leading cause of cancer-related deaths and the third most common cancer globally. Non-coding RNAs, including miRNAs, regulate the tumor microenvironment of CRC and play key roles in its progression. Abnormal levels of miR-150 are associated with cancer cell migration, invasion, and angiogenesis. The expression of miR-150 in fundamental biological processes is influenced by cancer cell expression profiles. Objectives: The present study aimed to estimate the level of miR-150-3p and miR-150-5p expression in CRC patients. Methods: The study involved 50 CRC patients and 50 control participants. Liver enzyme levels and renal functions were evaluated. miRNA was extracted from blood samples, followed by complementary DNA synthesis. The gene expression levels of miR-150-3p and miR-150-5p were measured. Results: The study found a significant increase in ALT and ALP levels in CRC patients, with highly significant differences of 0.01 and 0.001, respectively, while AST levels showed no significant difference between groups. Urea and creatinine levels also showed no significant differences. Gene expression analysis revealed that miR-150-3p levels were similar between patients and controls (non-significant fold change of 1.161), whereas miR-150-5p expression was reduced in CRC patients (fold change of 0.88). Conclusions: miR-150-5p is downregulated in CRC patients, highlighting its potential as a diagnostic biomarker. However, no significant changes were observed in miR-150-3p levels. The results may be affected by factors such as treatment protocols and disease stages.
Downloads
References
Chen X, Jia M, Ji J, Zhao Z, Zhao Y. Exosome-derived non-coding RNAs in the tumor microenvironment of colorectal cancer: Possible functions, mechanisms and clinical applications. Front Oncol. 2022;12:887532. doi: 10.3389/fonc.2022.887532. DOI: https://doi.org/10.3389/fonc.2022.887532
Zhang N, Hu X, Du Y, Du J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed Pharmacother. 2021;134:111099. doi: 10.1016/j.biopha.2020.111099. DOI: https://doi.org/10.1016/j.biopha.2020.111099
Schetter AJ, Okayama H, Harris CC. The Role of microRNAs in colorectal cancer. Cancer J. 2012;18(3). doi: 10.1097/PPO.0b013e318258b78f. DOI: https://doi.org/10.1097/PPO.0b013e318258b78f
Frixa T, Donzelli S, Blandino G. Oncogenic MicroRNAs: Key players in malignant transformation. Cancers (Basel). 2015;7(4):2466–2485. doi: 10.3390/cancers7040904. DOI: https://doi.org/10.3390/cancers7040904
Sur D, Burz C, Sabarimurugan S, Irimie A. Diagnostic and prognostic significance of miR-150 in colorectal cancer: A systematic review and meta-analysis. J Pers Med. 2020;10(3). doi: 10.3390/jpm10030099. DOI: https://doi.org/10.3390/jpm10030099
Bakhsh T, Alhazmi S, Farsi A, Yusuf AS, Alharthi A, Qahl SH, et al. Molecular detection of exosomal miRNAs of blood serum for prognosis of colorectal cancer. Sci Rep. 2024;14(1):8902. doi: 10.1038/s41598-024-58536-3. DOI: https://doi.org/10.1038/s41598-024-58536-3
Yadav V, Singh T, Sharma D, Garg VK, Chakraborty P, Ghatak S, et al. Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis. Cancers (Basel). 2024;16(18). doi: 10.3390/cancers16183183. DOI: https://doi.org/10.3390/cancers16183183
Mehrgou A, Ebadollahi S, Seidi K, Ayoubi-Joshaghani MH, Ahmadieh Yazdi A, Zare P, et al. Roles of miRNAs in colorectal cancer: Therapeutic implications and clinical opportunities. Adv Pharm Bull. 2021;11(2):233–247. doi: 10.34172/apb.2021.029. DOI: https://doi.org/10.34172/apb.2021.029
Xu SJ, Chen JH, Chang S, Li HL. The role of miRNAs in T helper cell development, activation, fate decisions and tumor immunity. Front Immunol. 2024;14. doi: 10.3389/fimmu.2023.1320305. DOI: https://doi.org/10.3389/fimmu.2023.1320305
Chandan K, Gupta M, Sarwat M. Role of host and pathogen-derived microRNAs in immune regulation during infectious and inflammatory diseases. Front Immunol. 2019;10:3081. doi: 10.3389/fimmu.2019.03081.
Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2). doi: 10.3390/cells9020276. DOI: https://doi.org/10.3390/cells9020276
Ameri A, Ahmed HM, Pecho RDC, Arabnozari H, Sarabadani H, Esbati R, et al. Diverse activity of miR-150 in Tumor development: shedding light on the potential mechanisms. Cancer Cell Int. 2023;23(1):261. doi: 10.1186/s12935-023-03105-3. DOI: https://doi.org/10.1186/s12935-023-03105-3
Balacescu O, Sur D, Cainap C, Visan S, Cruceriu D, Manzat-Saplacan R, et al. The impact of miRNA in colorectal cancer progression and its liver metastases. Int J Mol Sci. 2018;19(12). doi: 10.3390/ijms19123711. DOI: https://doi.org/10.3390/ijms19123711
Jin M, Yang Z, Ye W, Xu H, Hua X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS One. 2014;9(8):e103965. doi: 10.1371/journal.pone.0103965. DOI: https://doi.org/10.1371/journal.pone.0103965
Munshi SU, Panda H, Holla P, Rewari BB, Jameel S. MicroRNA-150 is a potential biomarker of HIV/AIDS disease progression and therapy. PLoS One. 2014;9(5):e95920. doi: 10.1371/journal.pone.0095920. DOI: https://doi.org/10.1371/journal.pone.0095920
He MM, Fang Z, Hang D, Wang F, Polychronidis G, Wang L, et al. Circulating liver function markers and colorectal cancer risk: A prospective cohort study in the UK Biobank. Int J cancer. 2021;148(8):1867–78. doi: 10.1002/ijc.33351. DOI: https://doi.org/10.1002/ijc.33351
Ben-Dov IZ, Muthukumar T, Morozov P, Mueller FB, Tuschl T, Suthanthiran M. microRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation. 2012;94(11):1086–1094. doi: 10.1097/TP.0b013e3182751efd. DOI: https://doi.org/10.1097/TP.0b013e3182751efd
Deng X, Lin Z, Zuo C, Fu Y. Upregulation of miR-150-5p alleviates LPS-induced inflammatory response and apoptosis of RAW264.7 macrophages by targeting Notch1. Open life Sci. 2020;15(1):544–52. doi: 10.1515/biol-2020-0058. DOI: https://doi.org/10.1515/biol-2020-0058
Ghareeb ZA, Al-Khafaji H, Al-Saedi MKA. Low-density lipoprotein receptor-related protein-1 and mir-205 expression for cardiovascular disease in familial hypercholesterolemia and non-familial hypercholesterolemia in Iraqi population. Iran J Ichthyol. 2021;8:246–254. DOI: https://doi.org/10.53293/jasn.2021.3691.1036
Wu Y, Huang A, Li T, Su X, Ding H, Li H, et al. MiR-152 reduces human umbilical vein endothelial cell proliferation and migration by targeting ADAM17. FEBS Lett. 2014;588(12):2063–2069. doi: 10.1016/j.febslet.2014.04.037. DOI: https://doi.org/10.1016/j.febslet.2014.04.037
Patel RK, West JD, Jiang Y, Fogarty EA, Grimson A. Robust partitioning of microRNA targets from downstream regulatory changes. Nucleic Acids Res. 2020;48(17):9724–9746. doi: 10.1093/nar/gkaa687. DOI: https://doi.org/10.1093/nar/gkaa687
Riedel G, Rüdrich U, Fekete-Drimusz N, Manns MP, Vondran FWR, Bock M. An extended ΔCT-method facilitating normalisation with multiple reference genes suited for quantitative RT-PCR analyses of human hepatocyte-like cells. PLoS One. 2014;9(3):e93031. doi: 10.1371/journal.pone.0093031. DOI: https://doi.org/10.1371/journal.pone.0093031
SPSS. Statistical Package for the Social Sciences. 2019. p. Version 26.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. DOI: https://doi.org/10.1038/nprot.2008.73
Martin J, Petrillo A, Smyth EC, Shaida N, Khwaja S, Cheow HK, et al. Colorectal liver metastases: Current management and future perspectives. World J Clin Oncol. 2020;11(10):761–808. doi: 10.5306/wjco.v11.i10.761. DOI: https://doi.org/10.5306/wjco.v11.i10.761
Miguel V, Ramos R, García-Bermejo L, Rodríguez-Puyol D, Lamas S. The program of renal fibrogenesis is controlled by microRNAs regulating oxidative metabolism. Redox Biol. 2021;40:101851. doi: 10.1016/j.redox.2020.101851. DOI: https://doi.org/10.1016/j.redox.2020.101851
Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol. 2009;15(45):5633–5640. doi: 10.3748/wjg.15.5633. DOI: https://doi.org/10.3748/wjg.15.5633
Kan Changez MI, Mubeen M, Zehra M, Samnani I, Abdul Rasool A, Mohan A, et al. Role of microRNA in non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH): a comprehensive review. J Int Med Res. 2023;51(9):3000605231197058. doi: 10.1177/03000605231197058. DOI: https://doi.org/10.1177/03000605231197058
Lee TH, Kim WR, Poterucha JJ. Evaluation of elevated liver enzymes. Clin Liver Dis. 2012;16(2):183–198. doi: 10.1016/j.cld.2012.03.006. DOI: https://doi.org/10.1016/j.cld.2012.03.006
Ray L, Nanda SK, Chatterjee A, Sarangi R, Ganguly S. A comparative study of serum aminotransferases in chronic kidney disease with and without end-stage renal disease: Need for new reference ranges. Int J Appl Basic Med Res. 2015;5(1):31–35. doi: 10.4103/2229-516X.149232. DOI: https://doi.org/10.4103/2229-516X.149232
Sette LHBC, Almeida Lopes EP de. Liver enzymes serum levels in patients with chronic kidney disease on hemodialysis: a comprehensive review. Clinics (Sao Paulo). 2014;69(4):271–278. doi: 10.4103/2229-516X.149232. DOI: https://doi.org/10.6061/clinics/2014(04)09
Yan S, Yim LY, Lu L, Lau CS, Chan VSF. MicroRNA regulation in systemic lupus erythematosus pathogenesis. Immune Netw. 2014;14(3):138–148. doi: 10.4110/in.2014.14.3.138. DOI: https://doi.org/10.4110/in.2014.14.3.138
Chandan K, Gupta M, Sarwat M. Role of host and pathogen-derived microRNAs in immune regulation during infectious and inflammatory diseases. Front Immunol. 2020;10:502685. doi: 10.3389/fimmu.2019.03081. DOI: https://doi.org/10.3389/fimmu.2019.03081
Zaravinos A. The regulatory role of microRNAs in EMT and cancer. J Oncol. 2015;2015:865816. doi: 10.1155/2015/865816. DOI: https://doi.org/10.1155/2015/865816
Min L, Zhu S, Chen L, Liu X, Wei R, Zhao L, et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles. 2019;8(1):1643670. doi: 10.1080/20013078.2019.1643670. DOI: https://doi.org/10.1080/20013078.2019.1643670
Zou SL, Chen YL, Ge ZZ, Qu YY, Cao Y, Kang ZX. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019;26(1):69–77. doi: 10.3233/CBM-190156. DOI: https://doi.org/10.3233/CBM-190156
Koshizuka K, Hanazawa T, Kikkawa N, Katada K, Okato A, Arai T, et al. Antitumor miR-150-5p and miR-150-3p inhibit cancer cell aggressiveness by targeting SPOCK1 in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2018;45(4):854–865. doi: 10.1016/j.anl.2017.11.019. DOI: https://doi.org/10.1016/j.anl.2017.11.019
Li S, Han W, He Q, Wang Y, Jin G, Zhang Y. Ginsenoside Rh2 suppresses colon cancer growth by targeting miRNA-150-3p /SRCIN1/Wnt axis. Acta Biochim Biophys Sin (Shanghai). 2023;55. doi: 10.3724/abbs.2023032. DOI: https://doi.org/10.3724/abbs.2023032
Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi: 10.1136/gutjnl-2011-301122. DOI: https://doi.org/10.1136/gutjnl-2011-301122
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin H, et al. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med. 2014;18(10):2125–2134. doi: 10.1111/jcmm.12398. DOI: https://doi.org/10.1111/jcmm.12398
He Z, Dang J, Song A, Cui X, Ma Z, Zhang Y. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed Pharmacother. 2020;121:109495. doi: 10.1016/j.biopha.2019.109495. DOI: https://doi.org/10.1016/j.biopha.2019.109495
Li C, Du X, Xia S, Chen L. MicroRNA-150 inhibits the proliferation and metastasis potential of colorectal cancer cells by targeting iASPP. Oncol Rep. 2018;40(1):252–260. doi: 10.3892/or.2018.6406. DOI: https://doi.org/10.3892/or.2018.6406
Mahmoudivar S, Zarredar H, Asadi M, Zafari V, Hashemzadeh S, Farzaneh R, et al. Serum miR-23 and miR-150 profiles as biomarkers for predicting recurrence following surgical intervention in colorectal cancer patients. Reports Biochem Mol Biol. 2024;12(4):540. doi: 10.61186/rbmb.12.4.540. DOI: https://doi.org/10.61186/rbmb.12.4.540
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Al-Rafidain Journal of Medical Sciences ( ISSN 2789-3219 )

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Published by Al-Rafidain University College. This is an open access journal issued under the CC BY-NC-SA 4.0 license (https://creativecommons.org/licenses/by-nc-sa/4.0/).











